Guidelines for droplet-based single-cell RNA-seq with fixed cells (10x Genomics Flex)
Due to the significant differences between the scRNA-Seq workflow from 10X genomics with fresh or fixed cells we decided to initially make you aware of the differences to provide some guidance. If you are already aware of this, feel free to directly jump to the guideline , as usual make sure you have carefully looked at the initial checklist before submitting samples.
ADVANTAGES AND DIFFERENCES OF 10X GENOMICS FLEX ASSAY TO 3’ SCRNA-SEQ WITH FRESH CELLS
Single-cell mRNA sequencing gives valuable information about the heterogeneity in gene expression present within a tissue or cell type. Such type of experiment critically depends on the quality of the cells - which can easily be affected not only by intrinsic properties of the tissue, but also by harsh dissociation methods, long collection times, and long waiting times from when cells are collected and loaded onto the system for encapsulation.
To overcome such limitations, an initial fixation step allows the preservation of the native transcriptome state of each cell until further processing takes place. This assay has three main advantages:
- Overcome quality constraints of highly sensitive cells1, as the fixation stops any kind of cell activity that might lead to degradation of transcripts or even cell death.
- Higher levels of flexibility both to the user and to the DcGC, in two main aspects:
- scheduling conflicts are avoided, because fixed cells can be safely stored for months until further processing, thus representing a significant advantage for experimental systems or experimental designs where sample collection has to be done outside normal working hours;
- batch effects in longitudinal assays are decreased, since many samples can be processed at the same time.
- Lower costs per cell, as an early sample barcoding step enables pooling of many samples into a single encapsulation reaction.
This guideline refers to the 10X Genomics Flex assay. The biggest difference between the Flex (fixed) and the Fresh (live) assays is the way that transcripts are captured and labelled. The standard assay can be considered an unbiased approach since mRNAs are captured by their poly-A tail and the molecule is amplified into a sequencing library. Sequencing information is directly derived from the mRNA molecule. The Flex assay, on the contrary, is targeted: capture of the mRNA relies on transcript-specific probes, which are themselves amplified into a sequencing library. Sequencing information thus derives from the probes instead of the actual transcripts.
Despite this crucial difference, both methods are essentially equivalent for discovery of subpopulations and differential expression analyses.
The probe panel is so far only available for mouse and human transcripts, thus the flex assay is still limited to these species. The panels cover virtually the entire transcriptome, with the exception of some highly variable transcripts. An overview of the covered transcripts can be found here .
It is possible to add custom probes to these panels, to target additional genes. In this case, special attention must be paid to the design of the probes; please find the requirements here.
BACKGROUND INFORMATION ON THE 10X GENOMICS FLEX ASSAY
The 10X Genomics Flex assay is designed as to enable a higher throughput per experiment, achieved through two sequential barcoding steps, a sample-specific barcode and a cell-specific barcode.
A Sample-specific barcode is introduced early in the protocol, when fixed cells|nuclei are incubated (in bulk) with a set of barcoded, transcript-specific probes which hybridize to their respective target mRNA molecules. This results in all transcripts from all cells within a sample being bound to probes with the same barcode. Cell-specific barcoding happens subsequently, when each cell gets encapsulated within a gel emulsion droplet (GEM) with a bead containing a unique, bead-specific barcode. The hybridized probes are captured be complementary sequences in the bead that also harbor the bead barcode, resulting – after amplification - in double-barcoded library molecules.
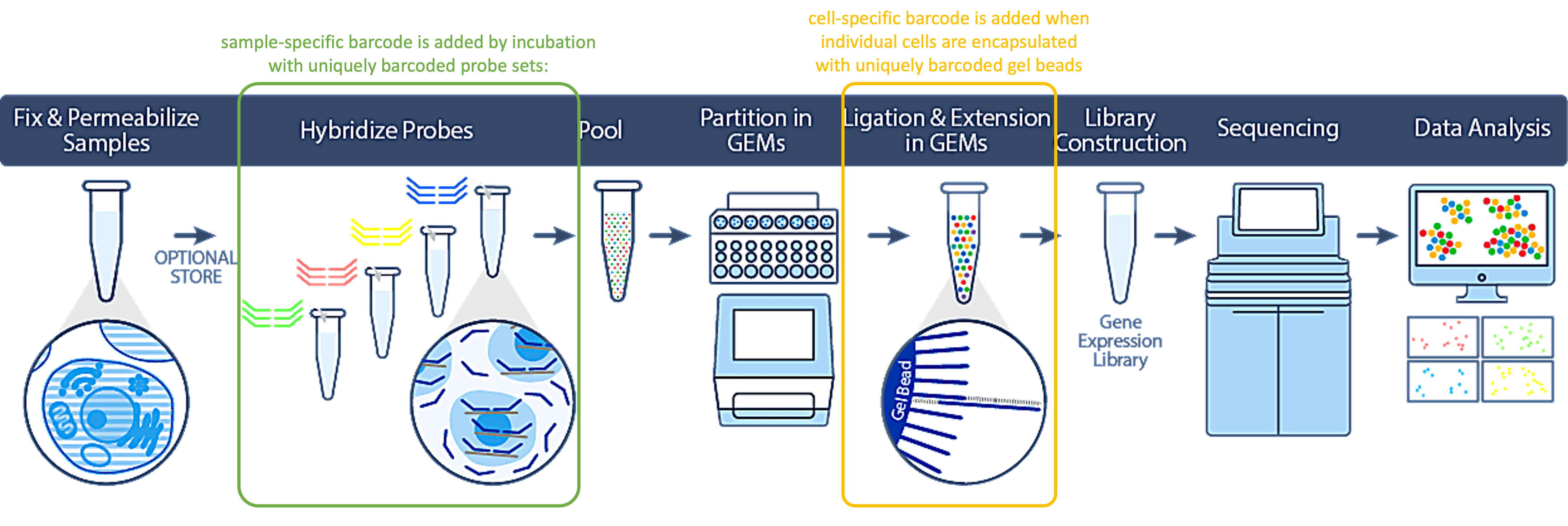
This dual-barcoding strategy allows a significant increase in experimental throughput via sample multiplexing. The kit allows for up to 16 different samples to be combined into one single reaction, for an output of 8.000 sequenced cells per sample. For optimal outcomes and optimal costs, we strongly recommend working with multiples of 4 samples. Currently, we offer multiplexing of 1, 4, 8, 12 or 16 samples.
Samples can be flexibly defined:
- each sample can come from e.g. a different individual, genetic background, experimental condition, or timepoint. In this case, around 8.000 (≥ 8 samples) or 10.000 cells (up to 4 samples) per sample are sequenced
- a sample can also come from one single condition which is then split into (up to) 16 sub-samples, if increased throughput is desired. In this example, up to 128.000 cells can be sequenced.
- ! ! ! We do not recommend pooling samples with different cell types or activity states together. If such approach is necessary, please be aware that it can result in an imbalanced sequencing coverage as cells with higher transcriptional activities will capture more sequencing reads.
Additional multiplexing is supported, by means of antibody-based labeling of the samples as a first step before fixation. Please contact us for further information.
Please get in touch to discuss your experimental design before actually planning the experiments.
GENERAL GUIDELINES FOR SAMPLE PREPARATION
CHECKLIST CRITICAL POINTS
FIXATION
Tissues (fresh, frozen or FFPE), cells, or nuclei can be used as input for the fixation step, to prepare samples for the 10X Genomics Flex assay. There are specific protocols for fixation of tissues (fresh and frozen or FFPE) and cells|nuclei. Please read them carefully before starting the protocol.
Tissue|cell|nuclei fixation must be done by the client. We are happy to support you in case of questions.
Fixed samples can be stored in -80°C for up to 6 months.
Accurate quantification of the number of cells in the samples is critical at this step. Please be aware that the sample with the lowest cell number will determine the recovery efficiency when pooling samples equally.
CELL QUALITY
A successful droplet-based single-cell|nuclei sequencing experiment in the 10x Chromium system is highly dependent on the quality of the cells. Therefore, it is important to be aware of some critical aspects of sample prep to ensure a proper and good preparation of the sample:
High cell viability rate before fixation: ideally greater than 90% of viable cells (minimum of 70%)
To increase the rate of viable cells in the cell suspension before fixation, we recommend to use the Miltenyi Dead Cell Removal Kit. FACS-sorting of living cells is also an option. Additionally, please check for cell viability with automated cell counters or manual count following trypan blue staining or equivalent.
Cell suspension should be clean of debris and aggregates
It is crucial that the cell suspension is clean of debris, and that cells or nuclei are not clumped together before fixation.
INPUT MATERIAL
Type & media
Fixed cells|nuclei in Enhancer (10X Genomics chemistry) with 50% glycerol, following the protocol for long-term storage at -80°C.
Amount & volume
As input for fixation, it is recommended to start with minimum 300.000 cells or 500.000 nuclei. The maximum is 10 million cells per sample. However, for optimal handling and results, we recommend starting with 1mio cells|nuclei.
SEQUENCING
Paired-end sequencing, 2x100 bp reads (different read lengths can be requested). Sequencing depth generally depends on cell type and research question. We recommend 12 k reads/cell or nuclei as a good starting point. In general, we observed that cells|nuclei derived from cell lines and organoids have a higher RNA content compared with those from primary culture. Therefore, we recommend a deeper sequencing of material from cultured cell lines and organoids.
Technical/biological replicates are dependent on experimental design and research question. We are glad to provide guidance with this.
BIOINFORMATICS ANALYSIS
Our single-cell RNA-seq analysis pipeline provides information about the number of cells that were sequenced, estimation of background noise, average number of genes expressed per cell|nuclei, clustering analysis, and a list of genes that characterizes each cluster. More information can be found here.
In case you need further support, or any other type of information, we can arrange a meeting with one of our bioinformatics experts to discuss the experiment.
In case you need further support, or any other type of information, we can arrange a meeting with one of our bioinformatics experts to discuss the experiment.
SAMPLE SUBMISSION
Important: Please prepare the fixed cells in 1.5 ml Eppendorf tubes. Tubes must be clearly labelled, both on the side wall and lid, using either a unique sample name or using the sample ID that is generated after entering the samples into our Rosalind client portal.
We strongly recommend using LoBind tubes or BSA-coated tubes for low cell numbers (<100 k cells in total). Use of non LoBind tubes will drastically reduce yields.
Providing project and sample information via the Rosalind Client Portal
Please log in to Rosalind to enter all necessary project information making use of the “Project Description” field during the project set-up dialogue. Give all relevant details about project design, sample preparation method and how the samples are eluted and/or preserved. Sample-specific information such as concentration, volume, used preparation protocols etc. should be provided when the samples are added to the created project.
Please also indicate if you require a bioinformatics analysis during project setup.
To submit samples, you must be registered in our Rosalind Portal. Therefore, please send an email to genomecenter@tu-dresden.de with your contact details (name, working group, working group leader, your email address and telephone number). This will enable us to set up an account for you. After you have received an automatic email, please log in to Rosalind with your login details. If needed, we can reset your login details.
Following up project progress
You can accompany the status of your project in our Rosalind Portal. Documents with information about the preparation method and quality control of samples and libraries are provided for your project.
REFERENCES
Footnotes
It is important to note that the success of such experiment still depends on good quality samples, thus we recommend to rigorously optimize collection procedures and remain very attentive to the quality and integrity of the cells.↩︎